ANGSD: Analysis of next generation Sequencing Data
Latest tar.gz version is (0.938/0.939 on github), see Change_log for changes, and download it here.
Abbababa: Difference between revisions
No edit summary |
No edit summary |
||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Performs the abbababa test also called the D-statistic. This tests for ancient admixture (or wrong tree topology). As all methods in ANGSD we require that the header of the BAM files are the same. | |||
; some of the options only works for the develupmental version availeble from github | |||
; if you use -rf to specify regions. These MUST appear in the same ordering as your fai file. | |||
__TOC__ | |||
<classdiagram type="dir:LR"> | <classdiagram type="dir:LR"> | ||
[ | [BAM files{bg:orange}]->[Sequence data|Random base] | ||
[sequence data]->[*.abbababa|ABBA and BABA couts file{bg:blue}] | [sequence data]->[*.abbababa|ABBA and BABA couts file{bg:blue}] | ||
</classdiagram> | </classdiagram> | ||
| Line 12: | Line 14: | ||
[*.abbababa|ABBA and BABA couts file{bg:blue}]->jackKnife.R[D stat and Z scores{bg:blue}] | [*.abbababa|ABBA and BABA couts file{bg:blue}]->jackKnife.R[D stat and Z scores{bg:blue}] | ||
</classdiagram> | </classdiagram> | ||
=Method= | |||
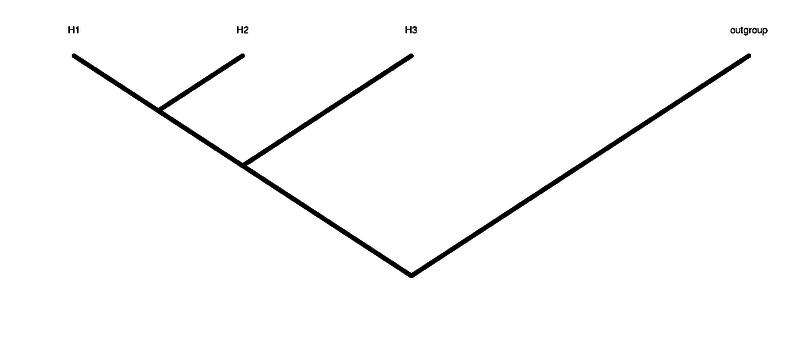
[[File:tree.png|800px]] | |||
=Brief Overview= | =Brief Overview= | ||
| Line 18: | Line 24: | ||
-------------- | -------------- | ||
abcDstat.cpp: | |||
-doAbbababa 0 | -doAbbababa 0 run the abbababa analysis | ||
-rmTrans 0 remove transitions | -rmTrans 0 remove transitions | ||
-blockSize 5000000 | -blockSize 5000000 size of each block in bases | ||
-enhance 0 outgroup must have same base for all reads | |||
-ans (null) fasta file with outgroup | |||
-useLast 0 use the last individuals as outgroup instead of -anc | |||
</pre> | </pre> | ||
| Line 29: | Line 36: | ||
=Options= | =Options= | ||
;- | ;-doAbbababa 1: sample a random base at each position. | ||
;-rmTrans [int] | |||
0; use all reads (default), 1 Remove transitions (important for ancient DNA) | |||
;-blockSize [INT] | |||
Size of each block. Choose a number that is higher than the LD in the populations. For human 5Mb (5000000) is usually used. | |||
; -anc [fileName.fa] | |||
Include an outgroup in fasta format. | |||
; -doCounts 1 | |||
use -doCounts 1 in order to count the bases at each sites after filters. | |||
; -useLast [int] | |||
1: use the last individual in the bam file list as outgroup instead of a fasta file (-anc) | |||
; -enhance [int] | |||
1: use only sites where the reads for the outgroup has the same base for all reads. Only works with -useLast 1 | |||
In order to do fancy filtering of bases based on quality scores see the [[Alleles_counts|Allele counts]] options. | |||
=Output= | =Output= | ||
Output | ;*.abbbababa | ||
Output: Each lines represents a block with a chromsome name (Column 1), a start position (Column 2), an end postion (Column 3). The new columns are the counts of ABBA and BABA sites. For each combination of 3 individuals (H1,H2,H3) two columns are printed. These number served as input to the R script called jackKnife.R. This script will skip combinations of individuals if there is less than 3 blocks with data. Type "Rscript R/jackKnife.R" to see additional options. | |||
=Jackknife= | |||
<pre> | |||
Rscript R/jackKnife.R | |||
-> needed arguments: | |||
file : the .abbababa filename | |||
indNames : list of individual names (you can use the bam.filelist) | |||
-> optional arguments (defaults): | |||
outfile ( out ) : name of output file | |||
boot ( 0 ) : print results for each bootstrap(jackknife), 0=NO | |||
</pre> | |||
==Example== | ==Example== | ||
| Line 41: | Line 77: | ||
<pre> | <pre> | ||
./angsd - | # select 5 individuals | ||
head -n5 bam.filelist > smallBam.filelist | |||
#run angsd | |||
./angsd -out out -doAbbababa 1 -bam smallBam.filelist -doCounts 1 -anc chimpHG19.fa | |||
#estimate Z score | |||
Rscript R/jackKnife.R file=out.abbababa indNames=smallBam.filelist outfile=out | |||
</pre> | |||
This results in a out.txt file with all the results. | |||
=Output= | |||
<pre> | |||
H1 H2 H3 nABBA nBABA Dstat jackEst SE Z | |||
NA11830 NA12004 NA12763 269 322 -0.08967851 -0.08967851 0.09006086 -0.9957545 | |||
NA11830 NA06985 NA12763 267 298 -0.05486726 -0.05486726 0.122256 -0.4487898 | |||
NA12004 NA06985 NA12763 254 243 0.0221328 0.0221328 0.1386198 0.1596655 | |||
NA11830 NA11993 NA12763 225 336 -0.197861 -0.197861 0.08514797 -2.323731 | |||
NA12004 NA11993 NA12763 217 267 -0.1033058 -0.1033058 0.09471542 -1.090697 | |||
NA06985 NA11993 NA12763 242 302 -0.1102941 -0.1102941 0.1241554 -0.8883553 | |||
NA12763 NA12004 NA11830 237 322 -0.1520572 -0.1520572 0.1047361 -1.451813 | |||
NA12763 NA06985 NA11830 219 298 -0.1528046 -0.1528046 0.1115283 -1.370098 | |||
</pre> | |||
'''H1 H2 H3''' are the 3 individuals in the tree that are not the outgroup. H1 and H2 are the ingroup (see figure of tree above) | |||
'''nABBA''' the total counts of ABBA patterns | |||
'''nBABA''' the total counts of BABA patterns | |||
'''Dstat''' The test statistic: (nABBA-nBABA)/(nABBA+nBABA). A negative value means that H1 is closer to H3 than H2 is. A positive value means that H2 is closer to H3 than H1 is. | |||
'''JackEst''' column is another estimate of the abbababa statistic that is bias corrected. This value is extremely similar to the value in the Dstat column | |||
'''SE''' is the estimated m-delete blocked Jackknife Standard error of the estimate used to obtain the Z value | |||
'''Z''' Z value that can be used to determine the significance of the test. As in Reich et al. an absolute value of the Z score above 3 is often used as a critical value. However, this note that this does not take into account the fact that we perform multiple tests. | |||
== bootstrap output == | |||
if you chose boot=1 then you will get an additional file out.boot. | |||
<pre> | |||
A06994 NA11840 NA06985 -0.03640257 -0.02449889 -0.03913043 -0.02643172 -0.004694836 -0.05909091 -0.007125891 | |||
NA06994 NA07056 NA06985 -0.04844291 0.001808318 -0.04727273 -0.1287879 -0.005671078 -0.04270463 -0.06569343 | |||
NA11840 NA07056 NA06985 -0.01075269 0.02592593 -0.003831418 -0.1062992 0.005847953 0.00952381 -0.05794393 | |||
NA06994 NA07357 NA06985 0.05856833 0.04587156 0.07798165 -0.02403846 0.07476636 0.06190476 0.05217391 | |||
NA11840 NA07357 NA06985 0.07014028 0.05714286 0.09401709 -0.02222222 0.05240175 0.09051724 0.04175824 | |||
NA07056 NA07357 NA06985 0.1285988 0.06530612 0.1361868 0.1322957 0.09913793 0.1216495 0.1422764 | |||
NA06985 NA11840 NA06994 -0.1950617 -0.1302211 -0.1802469 -0.1708543 -0.1002571 -0.1620948 -0.07614213 | |||
NA06985 NA07056 NA06994 -0.1139706 -0.08661417 -0.06273063 -0.07775769 -0.03100775 -0.1204589 -0.06764168 | |||
NA11840 NA07056 NA06994 0.007380074 0.009784736 0.06114398 0.02495202 0.01581028 -0.007905138 -0.03875969 | |||
NA06985 NA07357 NA06994 -0.02358491 0.01886792 0.04285714 0.006993007 0.04578313 0.02955665 0.00456621 | |||
NA11840 NA07357 NA06994 0.1530398 0.1478261 0.2227273 0.1629956 0.1382488 0.1748879 0.06451613 | |||
NA07056 NA07357 NA06994 0.1127542 0.1181102 0.1127542 0.1011236 0.09278351 0.1583166 0.0754352 | |||
NA06985 NA06994 NA11840 -0.1597938 -0.1060606 -0.1421189 -0.1450777 -0.09560724 -0.104 -0.06905371 | |||
NA06985 NA07056 NA11840 -0.2287582 -0.1662971 -0.1491228 -0.190678 -0.1697248 -0.1685393 -0.1915789 | |||
NA06994 NA07056 NA11840 -0.08467742 -0.05857741 -0.01061571 -0.06498952 -0.07559395 -0.06694561 -0.1428571 | |||
NA06985 NA07357 NA11840 -0.1571072 -0.1105769 -0.06801008 -0.1192214 -0.09319899 -0.1076115 -0.07389163 | |||
</pre> | </pre> | ||
The first 3 columns are the names of the individuals. The rest are the Dstat for each jacknife. Note that the number of bootstrapt can differ between tests since blocks without any data are discarded. | |||
Latest revision as of 10:46, 11 March 2016
Performs the abbababa test also called the D-statistic. This tests for ancient admixture (or wrong tree topology). As all methods in ANGSD we require that the header of the BAM files are the same.
- some of the options only works for the develupmental version availeble from github
- if you use -rf to specify regions. These MUST appear in the same ordering as your fai file.
<classdiagram type="dir:LR">
[BAM files{bg:orange}]->[Sequence data|Random base]
[sequence data]->[*.abbababa|ABBA and BABA couts file{bg:blue}] </classdiagram>
<classdiagram type="dir:LR">
[*.abbababa|ABBA and BABA couts file{bg:blue}]->jackKnife.R[D stat and Z scores{bg:blue}]
</classdiagram>
Method
Brief Overview
> ./angsd -doAbbababa -------------- abcDstat.cpp: -doAbbababa 0 run the abbababa analysis -rmTrans 0 remove transitions -blockSize 5000000 size of each block in bases -enhance 0 outgroup must have same base for all reads -ans (null) fasta file with outgroup -useLast 0 use the last individuals as outgroup instead of -anc
This function will counts the number of ABBA and BABA sites
Options
- -doAbbababa 1
- sample a random base at each position.
- -rmTrans [int]
0; use all reads (default), 1 Remove transitions (important for ancient DNA)
- -blockSize [INT]
Size of each block. Choose a number that is higher than the LD in the populations. For human 5Mb (5000000) is usually used.
- -anc [fileName.fa]
Include an outgroup in fasta format.
- -doCounts 1
use -doCounts 1 in order to count the bases at each sites after filters.
- -useLast [int]
1: use the last individual in the bam file list as outgroup instead of a fasta file (-anc)
- -enhance [int]
1: use only sites where the reads for the outgroup has the same base for all reads. Only works with -useLast 1
In order to do fancy filtering of bases based on quality scores see the Allele counts options.
Output
- .abbbababa
Output: Each lines represents a block with a chromsome name (Column 1), a start position (Column 2), an end postion (Column 3). The new columns are the counts of ABBA and BABA sites. For each combination of 3 individuals (H1,H2,H3) two columns are printed. These number served as input to the R script called jackKnife.R. This script will skip combinations of individuals if there is less than 3 blocks with data. Type "Rscript R/jackKnife.R" to see additional options.
Jackknife
Rscript R/jackKnife.R -> needed arguments: file : the .abbababa filename indNames : list of individual names (you can use the bam.filelist) -> optional arguments (defaults): outfile ( out ) : name of output file boot ( 0 ) : print results for each bootstrap(jackknife), 0=NO
Example
Create a fasta file bases from a random samples of bases.
# select 5 individuals head -n5 bam.filelist > smallBam.filelist #run angsd ./angsd -out out -doAbbababa 1 -bam smallBam.filelist -doCounts 1 -anc chimpHG19.fa #estimate Z score Rscript R/jackKnife.R file=out.abbababa indNames=smallBam.filelist outfile=out
This results in a out.txt file with all the results.
Output
H1 H2 H3 nABBA nBABA Dstat jackEst SE Z NA11830 NA12004 NA12763 269 322 -0.08967851 -0.08967851 0.09006086 -0.9957545 NA11830 NA06985 NA12763 267 298 -0.05486726 -0.05486726 0.122256 -0.4487898 NA12004 NA06985 NA12763 254 243 0.0221328 0.0221328 0.1386198 0.1596655 NA11830 NA11993 NA12763 225 336 -0.197861 -0.197861 0.08514797 -2.323731 NA12004 NA11993 NA12763 217 267 -0.1033058 -0.1033058 0.09471542 -1.090697 NA06985 NA11993 NA12763 242 302 -0.1102941 -0.1102941 0.1241554 -0.8883553 NA12763 NA12004 NA11830 237 322 -0.1520572 -0.1520572 0.1047361 -1.451813 NA12763 NA06985 NA11830 219 298 -0.1528046 -0.1528046 0.1115283 -1.370098
H1 H2 H3 are the 3 individuals in the tree that are not the outgroup. H1 and H2 are the ingroup (see figure of tree above)
nABBA the total counts of ABBA patterns
nBABA the total counts of BABA patterns
Dstat The test statistic: (nABBA-nBABA)/(nABBA+nBABA). A negative value means that H1 is closer to H3 than H2 is. A positive value means that H2 is closer to H3 than H1 is.
JackEst column is another estimate of the abbababa statistic that is bias corrected. This value is extremely similar to the value in the Dstat column
SE is the estimated m-delete blocked Jackknife Standard error of the estimate used to obtain the Z value
Z Z value that can be used to determine the significance of the test. As in Reich et al. an absolute value of the Z score above 3 is often used as a critical value. However, this note that this does not take into account the fact that we perform multiple tests.
bootstrap output
if you chose boot=1 then you will get an additional file out.boot.
A06994 NA11840 NA06985 -0.03640257 -0.02449889 -0.03913043 -0.02643172 -0.004694836 -0.05909091 -0.007125891 NA06994 NA07056 NA06985 -0.04844291 0.001808318 -0.04727273 -0.1287879 -0.005671078 -0.04270463 -0.06569343 NA11840 NA07056 NA06985 -0.01075269 0.02592593 -0.003831418 -0.1062992 0.005847953 0.00952381 -0.05794393 NA06994 NA07357 NA06985 0.05856833 0.04587156 0.07798165 -0.02403846 0.07476636 0.06190476 0.05217391 NA11840 NA07357 NA06985 0.07014028 0.05714286 0.09401709 -0.02222222 0.05240175 0.09051724 0.04175824 NA07056 NA07357 NA06985 0.1285988 0.06530612 0.1361868 0.1322957 0.09913793 0.1216495 0.1422764 NA06985 NA11840 NA06994 -0.1950617 -0.1302211 -0.1802469 -0.1708543 -0.1002571 -0.1620948 -0.07614213 NA06985 NA07056 NA06994 -0.1139706 -0.08661417 -0.06273063 -0.07775769 -0.03100775 -0.1204589 -0.06764168 NA11840 NA07056 NA06994 0.007380074 0.009784736 0.06114398 0.02495202 0.01581028 -0.007905138 -0.03875969 NA06985 NA07357 NA06994 -0.02358491 0.01886792 0.04285714 0.006993007 0.04578313 0.02955665 0.00456621 NA11840 NA07357 NA06994 0.1530398 0.1478261 0.2227273 0.1629956 0.1382488 0.1748879 0.06451613 NA07056 NA07357 NA06994 0.1127542 0.1181102 0.1127542 0.1011236 0.09278351 0.1583166 0.0754352 NA06985 NA06994 NA11840 -0.1597938 -0.1060606 -0.1421189 -0.1450777 -0.09560724 -0.104 -0.06905371 NA06985 NA07056 NA11840 -0.2287582 -0.1662971 -0.1491228 -0.190678 -0.1697248 -0.1685393 -0.1915789 NA06994 NA07056 NA11840 -0.08467742 -0.05857741 -0.01061571 -0.06498952 -0.07559395 -0.06694561 -0.1428571 NA06985 NA07357 NA11840 -0.1571072 -0.1105769 -0.06801008 -0.1192214 -0.09319899 -0.1076115 -0.07389163
The first 3 columns are the names of the individuals. The rest are the Dstat for each jacknife. Note that the number of bootstrapt can differ between tests since blocks without any data are discarded.