ANGSD: Analysis of next generation Sequencing Data
Latest tar.gz version is (0.938/0.939 on github), see Change_log for changes, and download it here.
Tutorial: Difference between revisions
| Line 128: | Line 128: | ||
[uppile] parsing 20 number of samples | [uppile] parsing 20 number of samples | ||
Change of chromo detected Waiting for nThreads:0 | Change of chromo detected Waiting for nThreads:0 | ||
->printing at chr: chr1 pos:3756580 chunknumber 500^CCaught SIGNAL: 2 will try to exit nicely (no more threads are created, we will wait for the current threads to finish) | ->printing at chr: chr1 pos:3756580 chunknumber 500 | ||
^CCaught SIGNAL: 2 will try to exit nicely (no more threads are created, we will wait for the current threads to finish) | |||
SEnding NULL this is a killswitch | SEnding NULL this is a killswitch | ||
-> Done reading data waiting for calculations to finish | -> Done reading data waiting for calculations to finish | ||
Revision as of 08:56, 3 May 2013
This tutorial only uses BAMfiles.
Prepare files
angsd can work on remote bam files therefore first download a list with 23 unrelated europeans from the 1000genomes project.
wget http://popgen.dk/netstuff/files.list
Contents of the file 'files.list'
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12003/alignment/NA12003.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12004/alignment/NA12004.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12005/alignment/NA12005.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12006/alignment/NA12006.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11830/alignment/NA11830.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11831/alignment/NA11831.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11832/alignment/NA11832.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11992/alignment/NA11992.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11993/alignment/NA11993.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11994/alignment/NA11994.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11995/alignment/NA11995.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12154/alignment/NA12154.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12155/alignment/NA12155.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12156/alignment/NA12156.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA06994/alignment/NA06994.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11840/alignment/NA11840.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12044/alignment/NA12044.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12813/alignment/NA12813.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12760/alignment/NA12760.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12762/alignment/NA12762.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA06985/alignment/NA06985.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA11881/alignment/NA11881.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/NA12249/alignment/NA12249.mapped.ILLUMINA.bwa.CEU.low_coverage.20120522.bam
Understaing angsd options
As a simple reference for the program we have made most of the methods within angsd easy viewable by writing the associated command. All options are given by
-parameter value
It's important that there are no space between the dash and the paramater, it is important that there are a space betwwen the parameter and the value. Futhermore the parameter is casesensitive.
Simply writing angsd will give you the helpscreen.
./angsd
Command: angsd -> angsd version: 0.515 build(Mar 23 2013 12:35:04) -> Please use the website "http://www.popgen.dk/angsd" as reference -> Use -nThreads for number of threads allocated to the program Overview of methods: -GL estimate genotype likelihoods -doCounts Calculate various counts statistics -doDepth Do depth statistics -doAsso perform association study -doMaf estimate allele frequencies -doError estimate the type specific error rates -doAnsError estimate the errorrate based on perfect fastas -doHWE Est inbreedning per site -doGeno call genotypes -realSFS Estimate the SFS and/or perform neutrality tests Below are options that can be usefull -bam Options relating to bam reading -doMajorMinor Infer the major/minor using different approaches -ref/-anc Read reference or ancestral genome many others For information of specific options type: ./angsd METHODNAME eg ./angsd -GL ./angsd -doMaf ./angsd -doAsso etc Examples: Estimate MAF for bam files in 'list' './angsd -bam list -GL 2 -doMaf 2 -out RES -doMajorMinor 1'
An explanation for every parameter is shown beside the parameter, and for every of these options we can get additional information by typing that parameter solely without any options. An example below for the methods relating to genotype likelihood calculation.
./angsd -GL
-> angsd version: 0.515 build(Mar 27 2013 11:15:58) -> Analysis helpbox/synopsis information: --------------------- analysisEstLikes.cpp: -GL=0: 1: SAMtools 2: GATK 3: SOAPsnp 4: SYK -minQ 13 (remove bases with qscore<minQ) -trim 0 (zero means no trimming) -tmpdir angsd_tmpdir/ (used by SOAPsnp) -errors (null) (used by SYK) -minInd -1 (-1 indicates no filtering) Filedumping: -doGlf=0 1: binary glf (10 log likes) .glf 2: beagle likelihood file .beagle.gz 3: binary 3 times likelihood .glf 4: text version (10 log likes) .glf
This tells you that there are 4 different genotype likelihood models implemented and you can choose accordingly by writing -GL 1 for the SAMtools model. We also see that we can dump the genotype likelihoods in four different ways.
Understanding angsd output
Program catches system signals, if you press ctrl+c, it will therefore stop the filereading, but will let the threads already running finish their jobs. You can therefore press ctrl+c at anytime at expect to get proper output files. After a run has been completed the program will printout a list of the generated files.
An example is below.
./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf -doMajorMinor 1 -GL 1 -nInd 20 -minMaf 0.005 -nThreads 10 -doCounts 1 -dumpCounts 3
Command: ./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf -doMajorMinor 1 -GL 1 -nInd 20 -minMaf 0.005 -nThreads 10 -doCounts 1 -dumpCounts 3 -> angsd version: 0.529 build(May 2 2013 14:09:03) ->Starting analysis [uppile] parsing 20 number of samples Change of chromo detected Waiting for nThreads:0 ->printing at chr: chr1 pos:3756580 chunknumber 500 ^CCaught SIGNAL: 2 will try to exit nicely (no more threads are created, we will wait for the current threads to finish) SEnding NULL this is a killswitch -> Done reading data waiting for calculations to finish -> Calling destroy -> Waiting for nThreads:9 -> Waiting for nThreads:3 -> Done waiting for threads ->Output filenames: ->"tstMaf.arg" ->"tstMaf.pos" ->"tstMaf.counts" ->"tstMaf.mafs" Thu May 2 15:11:31 2013 [ALL done] cpu-time used = 77.95 sec [ALL done] walltime used = 21.00 sec
Getting simple Counts/depth
For some analysis simply getting the sequencing depth for all sites could be of interest, this can of analysis is grouped in the '-doCounts' methods.
./angsd -doCounts
--------------- analysisCount.cpp: -doCounts 0 (Count the number A,C,G,T. All sites, All samples) -minQ 13 (remove bases with qscore<minQ) -setMaxDepth -1 (-1 indicates no filtering) -trim 0 (trim ends of reads) -minInd 0 (0 indicates no filtering) Filedumping: -doDepth 0 (dump distribution of seqdepth) .depthSample,.depthGlobal -maxDepth 100 (bin together high depths) -doQsDist 0 (dump distribution of qscores) .qs -dumpCounts 0 1: total seqdepth for site .pos 2: seqdepth persample .pos,.counts 3: A,C,G,T overall all samples .pos,.counts 4: A,C,G,T for all samples .pos,.counts
So if we wanted the sum of ACGTS across all samples we could write
./angsd -bam files.list -doCounts 1 -dumpCounts 3 -out tstCounts -nInd 10
--------------- head tstCounts.counts totA totC totG toft 0 0 1 0 0 1 0 0 1 0 0 0 1 0 0 0 2 0 0 0 0 0 0 2 0 0 0 2 0 0 0 2 0 0 2 0
Or if we wanted the sequencing depth per sample but only for the good quality data
./angsd -bam files.list -doCounts 1 -dumpCounts 2 -out tstCounts -nInd 10 -minQ 20 -minMapQ 30
--------------- head tstCounts.counts ind0 ind1 ind2 ind3 ind4 ind5 ind6 ind7 ind8 ind9 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0
Frequencies
We can also estimate the allele frequencies. This we do by using the -doMaf option.
./angsd -doMaf
analysisMaf.cpp: -doMaf 0 1: BFGS frequency (known major minor) 2: EM frequency (known major minor) 4: BFGS frequency (unknown major minor) 8: EM frequency (unknown major minor) 16: frequency from genotype probabilities 32: alleleCounts based method (known major minor) -doSNP 0 -minMaf 0.010000 0 -minLRT 24.000000 0 -ref (null) -anc (null) -doZ 0 -eps 0.001000 [Only used for -doMaf &32]
So if we try to use -doMaf 2 angsd will complain!
./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf
Error: you need to specify doMajorMinor in order to doMaf
So lets decide also estimate the major and minor, what are the options.
./angsd -doMajorMinor
------------------- analysisMajorMinor.cpp: -doMajorMinor 0 1: Infer major and minor from GL 2: Infer major and minor from allele counts 3: use major and minor from bim file (requires -filter afile.bim) 4: Use reference allele as major (requires -ref) 5: Use ancestral allele as major (requires -anc)
Let us infer the major and minor using the genotype likelihoods.
./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf -doMajorMinor 1
-doMajorMinor 1 is based on genotype likelihoods, you must specify a genotype likelihood model -GL
So now we need to specify which genotype likelihood model we want to use, let us see what our options are
./angsd0.530/angsd -GL
--------------------- analysisEstLikes.cpp: -GL=0: 1: SAMtools 2: GATK 3: SOAPsnp 4: SYK -minQ 13 (remove bases with qscore<minQ) -trim 0 (zero means no trimming) -tmpdir angsd_tmpdir/ (used by SOAPsnp) -errors (null) (used by SYK) -minInd -1 (-1 indicates no filtering) Filedumping: -doGlf 0 1: binary glf (10 log likes) .glf 2: beagle likelihood file .beagle.gz 3: binary 3 times likelihood .glf 4: text version (10 log likes) .glf
We pick the same model they use in samtools '-GL 1'.
./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf -doMajorMinor 1 -GL 1 -nInd 20
head tstMaf.mafs chromo position major minor knownEM nInd chr1 11782 G A 0.000008 1 chr1 11783 C A 0.000008 1 chr1 11784 A C 0.000008 1 chr1 11785 A C 0.000008 2 chr1 11786 A C 0.000008 3 chr1 11787 T A 0.000008 3 chr1 11788 T A 0.000008 3 chr1 11789 T A 0.000008 4 chr1 11790 G A 0.000008 4
These sites are all invariable so lets filter out the sites with a maf below 0.5%
./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf -doMajorMinor 1 -GL 1 -nInd 20 -minMaf 0.005 -nThreads 10
head tstMaf.mafs chromo position major minor knownEM nInd chr1 13032 A T 0.016541 9 chr1 13038 T C 0.073424 9 chr1 13309 G T 0.207854 4 chr1 13396 T A 0.012727 16 chr1 13482 G C 0.019104 15 chr1 13502 G A 0.025732 14 chr1 13519 T C 0.018256 17 chr1 14933 A G 0.487768 1 chr1 16259 T G 0.060360 7
The filters works across the different analysis classes, so if we supply the dumpCounts we will only get the sites with a maf >0.5%
./angsd0.530/angsd -bam files.list -doMaf 2 -out tstMaf -doMajorMinor 1 -GL 1 -nInd 20 -minMaf 0.005 -nThreads 10 -doCounts 1 -dumpCounts 3
paste tstMaf.pos tstMaf.counts |head chr pos totDepth totA totC totG totT chr1 13032 21 20 0 0 1 chr1 13038 21 0 1 0 20 chr1 13309 8 0 0 7 1 chr1 13396 34 1 0 0 33 chr1 13482 30 0 1 29 0 chr1 13502 25 1 0 24 0 chr1 13519 26 0 1 0 25 chr1 14933 2 1 0 1 0 chr1 16259 9 0 0 1 8
Estimating the SFS
We can also use angsd for estimating the site frequency spectrum.
./angsd0.530/angsd -realSFS
-------------- angsd_realSFS.cpp: -realSFS 0 1: perform multisample GL estimation 2: use an inbreeding version -doThetas 0 (calculate thetas) -underFlowProtect 0 -fold 0 (deprecated) -anc (null) (ancestral fasta) -noTrans 0 (remove transitions) -pest (null) (prior SFS)
We have butterfly dataset for which we want to find the SFS.
So let us try to estimate the sfs
.angsd0.530/angsd -bam bom.bam -realSFS 1 -out testSFS
-------------- -> angsd version: 0.529 build(May 2 2013 14:09:03) Must supply -anc for polarizing the spectrum
So we also need to supply a fasta which should contain our ancestral states. So lets do that.
./angsd0.530/angsd -bam bom.bam -realSFS 1 -out testSFS -anc bombyx/input/referenceseq.fasta -r ref_contig:-100000
-------------- Command: angsd0.530/angsd -bam bom.bam -realSFS 1 -out testSFS -anc bombyx/input/referenceseq.fasta -r ref_contig:-100000 -> angsd version: 0.529 build(May 2 2013 14:09:03) Must supply genotype likelihoods (-GL [INT])
So we also need to pick a genotype likelihood model
angsd0.530/angsd -bam bom.bam -realSFS 1 -out testSFS -anc bombyx/input/referenceseq.fasta -r ref_contig:-100000 -GL 1
-------------- -> angsd version: 0.529 build(May 2 2013 14:09:03) [getFasta.cpp.init:24] bombyx/input/referenceseq.fasta ->Starting analysis [uppile] parsing 20 number of samples region lookup 1/1 Change of chromo detected Waiting for nThreads:0 [magic] chaning to chr: 0 ->printing at chr: ref_contig pos:91269 chunknumber 100SEnding NULL this is a killswitch -> Done reading data waiting for calculations to finish -> Calling destroy -> Done waiting for threads ->Output filenames: ->"testSFS.arg" ->"testSFS.sfs" ->"testSFS.sfs.pos" Thu May 2 16:57:57 2013 [ALL done] cpu-time used = 4.45 sec [ALL done] walltime used = 5.00 sec
We know use the external angsd program, for finding the MLE of the sfs
angsd0.530/misc/emOptim
-> -binput -nChr -maxIter -nThread -outnames
we need to supply the binary .sfs file and the number of chromosomes.
angsd0.530/misc/emOptim -binput testSFS.sfs -nChr 40 -nThread 2
-> Thu May 2 17:03:03 2013 dumped:testSFS.sfs.em testSFS.sfs.em.ml testSFS.sfs.em.log
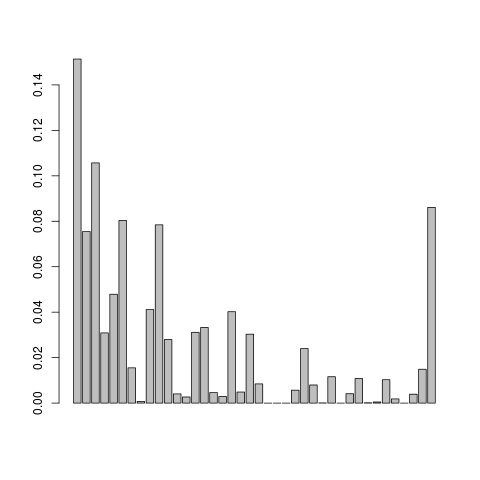
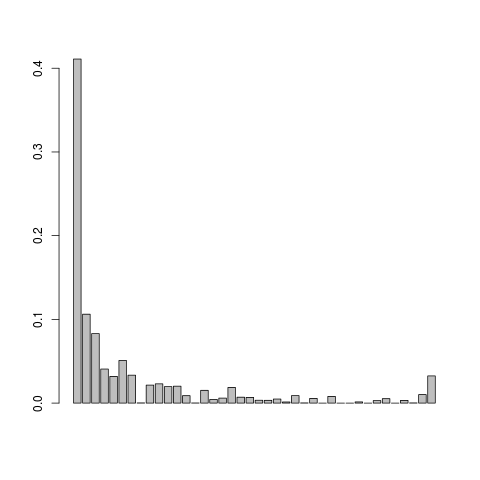
Let us visualise the .em.ml file
This doesn't look so nice, we can try adjusting the mapping quality and do local realignment of the reads