CATS: Difference between revisions
Jump to navigation
Jump to search
| Line 12: | Line 12: | ||
= plot examples = | = plot examples = | ||
==== | ==== Which design is better ==== | ||
[[File:catsFig1.png|thumb]] | [[File:catsFig1.png|thumb]] | ||
<pre> | <pre> | ||
library(CATS) | library(CATS) | ||
rr<-seq(1,2,by=0.05) | |||
maf<-c(0.05,0.1,0.2,0.5) | |||
c2<-curve.cats(rr,maf,ncases=600,ncontrols=600,ncases2=600,ncontrols2=600,alpha=0.000001,prevalence=0.01); | |||
plot(c2,type="One",main="power2",ylab="Power",xlab="RR",file=NULL,col=1:4) | |||
lines.cats(c2,type="Replication",lty=3) | |||
lines.cats(c2,type="Joint",lty=2) | |||
lines.cats(c2,type="First",lty=4) | |||
legend("left",c("One stage","Joint","Relication","First Stage"),lty=1:4,bty="n") | |||
</pre> | |||
==== Number of SNPs for replication ==== | |||
[[File:catsFig2.png|thumb]] | |||
<pre> | |||
library(CATS) | |||
power.J96<-c() | |||
power.J1536<-c() | |||
RR<-seq(1.1,1.5,by=0.025) | |||
maf=c(5,10,20,50)/100 | |||
for(tal2 in 1:length(maf)){ | |||
J1<-c() | |||
J2<-c() | |||
for(tal in 1:length(RR)){ | |||
temp<-cats(freq=maf[tal2],ncases=1500,ncontrols=1500,ncases2=2000,ncontrols2=2000,risk=RR[tal],pimarkers = 96/300000,alpha=0.05/300000) | |||
J1[tal]<-temp$P.joint | |||
temp<-cats(freq=maf[tal2],ncases=1500,ncontrols=1500,ncases2=2000,ncontrols2=2000,risk=RR[tal],pimarkers = 1536/300000,alpha=0.05/300000) | |||
J2[tal]<-temp$P.joint | |||
} | |||
power.J96<-cbind(power.J96,J1) | |||
power.J1536<-cbind(power.J1536,J2) | |||
} | |||
col=1:length(maf) | |||
plot(RR,power.J1536[,1],type=type,lwd=2,ylab="Power",main="Multiplicative model, 1500:1500, 2000:2000",col=col[1],ylim=0:1) | |||
for(tal2 in 2:length(maf)){ | |||
lines(RR,power.J1536[,tal2],lwd=2,col=col[tal2],type=type) | |||
} | |||
for(tal2 in 1:length(maf)){ | |||
lines(RR,power.J96[,tal2],lwd=2,col=col[tal2],type=type,lty=2) | |||
} | |||
legend("bottomright",c(paste("MAF=",c(maf),", rep=1536"),paste("MAF=",c(maf),", rep=96")),col=col,lwd=2,bty="n",lty=c(rep(1,length(maf)),rep(2,length(maf)))) | |||
</pre> | |||
==== heatmap of power ==== | |||
[[File:catsFig3.png|thumb]] | |||
<pre> | |||
library(CATS) | |||
source("superCATS.R") | |||
rr<-seq(1,2,by=0.025) | |||
c<-super.cats(rr,by=length(rr),ncases=765,ncontrols=1274,ncases2=100,ncontrols2=100,alpha=0.001,prevalence=0.01); | |||
bitmap("PowerFig3.png") | |||
plot(c,main="power",file=NULL) | |||
</pre> | |||
==== design and MAF ==== | |||
[[File:catsFig4.png|thumb]] | |||
<pre> | |||
library(CATS) | |||
source("superCATS.R") | |||
rr<-seq(1,2,by=0.05) | rr<-seq(1,2,by=0.05) | ||
maf<-c(0.05,0.1,0.2,0.5) | maf<-c(0.05,0.1,0.2,0.5) | ||
Revision as of 14:50, 5 August 2013
joint power of a non-symetric two stage GWA design
R package for power estimation for a two-stage genome-wide association design. This is a modification of the code from Skol et al 2006, nat genet. so that the relative risk, case-control ratios and allele frequencies are allowed to vary between stages.
Download and installation
plot examples
Which design is better
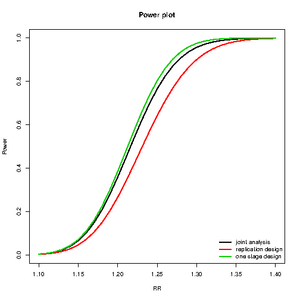
library(CATS)
rr<-seq(1,2,by=0.05)
maf<-c(0.05,0.1,0.2,0.5)
c2<-curve.cats(rr,maf,ncases=600,ncontrols=600,ncases2=600,ncontrols2=600,alpha=0.000001,prevalence=0.01);
plot(c2,type="One",main="power2",ylab="Power",xlab="RR",file=NULL,col=1:4)
lines.cats(c2,type="Replication",lty=3)
lines.cats(c2,type="Joint",lty=2)
lines.cats(c2,type="First",lty=4)
legend("left",c("One stage","Joint","Relication","First Stage"),lty=1:4,bty="n")
Number of SNPs for replication
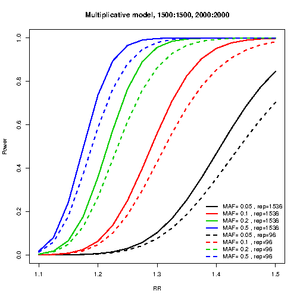
library(CATS)
power.J96<-c()
power.J1536<-c()
RR<-seq(1.1,1.5,by=0.025)
maf=c(5,10,20,50)/100
for(tal2 in 1:length(maf)){
J1<-c()
J2<-c()
for(tal in 1:length(RR)){
temp<-cats(freq=maf[tal2],ncases=1500,ncontrols=1500,ncases2=2000,ncontrols2=2000,risk=RR[tal],pimarkers = 96/300000,alpha=0.05/300000)
J1[tal]<-temp$P.joint
temp<-cats(freq=maf[tal2],ncases=1500,ncontrols=1500,ncases2=2000,ncontrols2=2000,risk=RR[tal],pimarkers = 1536/300000,alpha=0.05/300000)
J2[tal]<-temp$P.joint
}
power.J96<-cbind(power.J96,J1)
power.J1536<-cbind(power.J1536,J2)
}
col=1:length(maf)
plot(RR,power.J1536[,1],type=type,lwd=2,ylab="Power",main="Multiplicative model, 1500:1500, 2000:2000",col=col[1],ylim=0:1)
for(tal2 in 2:length(maf)){
lines(RR,power.J1536[,tal2],lwd=2,col=col[tal2],type=type)
}
for(tal2 in 1:length(maf)){
lines(RR,power.J96[,tal2],lwd=2,col=col[tal2],type=type,lty=2)
}
legend("bottomright",c(paste("MAF=",c(maf),", rep=1536"),paste("MAF=",c(maf),", rep=96")),col=col,lwd=2,bty="n",lty=c(rep(1,length(maf)),rep(2,length(maf))))
heatmap of power
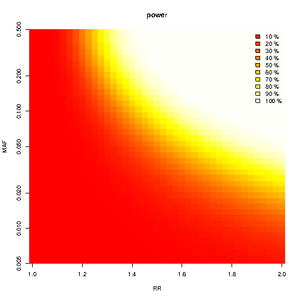
library(CATS)
source("superCATS.R")
rr<-seq(1,2,by=0.025)
c<-super.cats(rr,by=length(rr),ncases=765,ncontrols=1274,ncases2=100,ncontrols2=100,alpha=0.001,prevalence=0.01);
bitmap("PowerFig3.png")
plot(c,main="power",file=NULL)
design and MAF
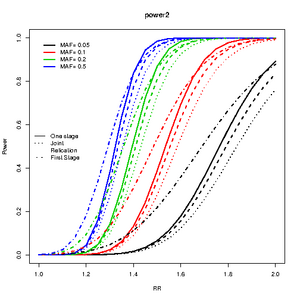
library(CATS)
source("superCATS.R")
rr<-seq(1,2,by=0.05)
maf<-c(0.05,0.1,0.2,0.5)
c2<-curve.cats(rr,maf,ncases=600,ncontrols=600,ncases2=600,ncontrols2=600,alpha=0.000001,prevalence=0.01);
plot(c2,type="One",main="power2",ylab="Power",xlab="RR",file=NULL,col=1:4)
lines.cats(c2,type="Replication",lty=3)
lines.cats(c2,type="Joint",lty=2)
lines.cats(c2,type="First",lty=4)
legend("left",c("One stage","Joint","Relication","First Stage"),lty=1:4,bty="n")