Main Page: Difference between revisions
| Line 18: | Line 18: | ||
= [[fastNGSadmix]] = | = [[fastNGSadmix]] = | ||
Infers the ancestry proportions from NGS data of a single indivudal, even with low-depth NGS data. It is based on ngsAdmix, however it uses a referenece panel of population specific frequencies, and works when you have uncertainty in your data. This makes it ideal for quick and easy analyses of medium and low depth sequencing data samples where many genotypes cannot be called without introducing errors or ascertainment bias. There is also a PCA method for doing PCA of a NGS sample, also taking population structure into account. | |||
= [[Relate]]= | = [[Relate]]= | ||
Revision as of 16:05, 5 March 2017
ANGSD
Analysis of Next Generation Sequencing Data
<classdiagram type="dir:LR"> [sequence data]->[genotype;likelihoods] [genotype;likelihoods]->[genotype;probabilities] [sequence files|bam files;SOAP files{bg:orange}]->[sequence data] [glf files|glfv3;soapSNP{bg:orange}]->[genotype;likelihoods] [genotype prob|beagle output{bg:orange}]->[genotype;probabilities] </classdiagram>
NgsAdmix
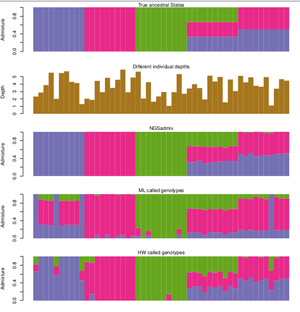
Infer the ancestry proportions from low depth NGS data. The principal is the same as other softwares such as FRAPPE and ADMIXTURE however, ngsAdmix also works when you have uncertainty in your data. This makes it ideal for medium and low depth sequencing data where many genotypes cannot be called without introducing errors or ascertainment bias.
fastNGSadmix
Infers the ancestry proportions from NGS data of a single indivudal, even with low-depth NGS data. It is based on ngsAdmix, however it uses a referenece panel of population specific frequencies, and works when you have uncertainty in your data. This makes it ideal for quick and easy analyses of medium and low depth sequencing data samples where many genotypes cannot be called without introducing errors or ascertainment bias. There is also a PCA method for doing PCA of a NGS sample, also taking population structure into account.
Relate
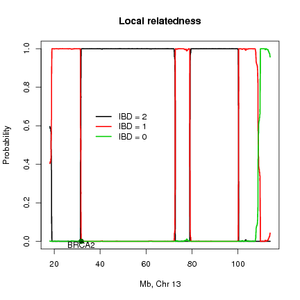
This method estimates the probability of sharing alleles identity by descent (IBD) across the genome and can also be used for mapping disease loci using distantly related individuals. To accommodate LD the methods need SNP for several individuals in order to estimate the allele frequencies and the pairwise LD. The method return the posterior probabilities of the IBD states across the genome and the overall IBD sharing.
RelateAdmix
A software package for inferring relatedness between pairs of individuals even if the individuals are admixed.
NgsRelate
A software package for inferring relatedness between pairs of individuals from NGS data.
BAMSE
CATS
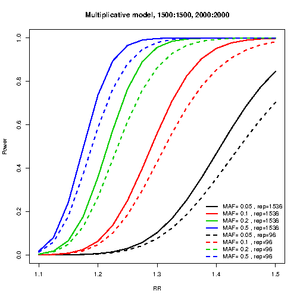
R package for power estimation for a two-stage genome-wide association design. This is a modification of the code from Skol et al 2006, nat genet. so that the relative risk, case-control ratios and allele frequencies are allowed to vary between stages.